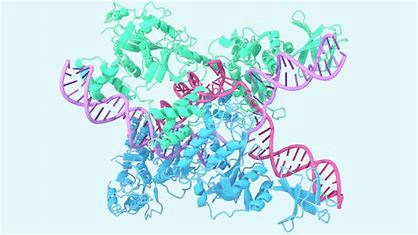
Proteins are essential molecules for life, playing a crucial role in numerous biological processes. They are present in all living cells and perform a multitude of vital functions. Composed of amino acids, they fold into specific three-dimensional structures that determine their functions.
These complex structures enable proteins to interact with other molecules, catalyze chemical reactions, transmit cellular signals, and provide structural support to cells and tissues.
However, predicting the exact structure of a protein from its amino acid sequence has long been a significant challenge in biology and biochemistry. Understanding this structure is essential, as it facilitates better insight into the mechanisms of protein action and the development of strategies to modulate their function, which is crucial for creating new drugs and treatments.
It is in this context that AlphaFold stands out as a revolutionary breakthrough in the field of biology.
What is AlphaFold?
AlphaFold is an artificial intelligence (AI) program created by DeepMind, a Google subsidiary specializing in deep learning. AlphaFold uses neural networks to accurately predict the three-dimensional structure of proteins from their amino acid sequences. This innovation has the potential to transform our understanding of fundamental biological processes and accelerate advances in medicine and biotechnology.
The challenges of protein structure prediction
Predicting protein structures represents a considerable challenge in molecular biology due to several complex factors.
1. Diversity of sequences and structures:
To date, more than 200 million proteins are known, with more discovered each year. Each has a unique three-dimensional shape.
Indeed, proteins are composed of 20 different types of amino acids, arranged in sequences that vary in length and composition. This diversity generates a multitude of possible three-dimensional structures, making accurate prediction extremely difficult.
2. Limits of experimental methods:
Various experimental methods such as X-ray crystallography or nuclear magnetic resonance (NMR) are used to determine protein structures. These methods, however, are time-consuming, costly, and not always successful.
Moreover, some proteins are challenging, if not impossible, to obtain precise structural data for using traditional experimental methods. These include very large, very flexible, or those that do not easily crystallize.
This is why, for decades, scientists have sought a method to reliably determine a protein’s structure from its amino acid sequence alone.

The success of AlphaFold
The CASP (Critical Assessment of Structure Prediction) competition is an event held every two years to evaluate methods for predicting protein three-dimensional structures.
For this purpose, newly experimentally determined protein structures (but not yet published) are selected as targets. In the following weeks, participating teams must predict these protein structures using their methods. Then, the predictions are compared to the actual experimental structures to assess the accuracy of the different prediction methods.
In 2018, DeepMind participated for the first time. From this session (CASP13), AlphaFold proved to be more efficient than all its competitors.
During CASP14 in 2020, AlphaFold outperformed all other teams with unprecedented accuracy, achieving levels comparable to traditional experimental methods. This success was hailed as a major breakthrough in the field.
How does AlphaFold work?
AlphaFold uses a combination of deep learning techniques and structural modeling to predict protein structures. Here are the main steps of the process:
- Data Input: The linear sequence of amino acids of the target protein is provided. AlphaFold generates multiple sequence alignments (MSA) to find similar sequences in protein databases, providing evolutionary information.
- Modeling: AlphaFold uses deep learning models, including transformers, to analyze relationships between amino acids. Transformers can handle long-distance relationships in sequences, crucial for predicting interactions between residues that are distant in the linear sequence but close in the 3D structure.
- Prediction of Distances and Angles: AlphaFold predicts distances between pairs of amino acids and angles of chemical bonds, helping to determine the protein’s 3D shape.
- Structural Assembly: Using distance and angle predictions, AlphaFold assembles the protein’s three-dimensional structure by minimizing an energy function that penalizes unrealistic configurations.
- Prediction Evaluation: The predicted structure is assessed for accuracy against available experimental data, and refinement techniques are used to improve the model’s quality.

Applications of AlphaFold
By enabling rapid and accurate prediction of protein structures, AlphaFold opens new avenues for biomedical and pharmaceutical research. For example:
- Drug Development: Knowledge of protein structures facilitates the design of drugs targeting specific proteins involved in diseases.
- Synthetic Biology: Scientists can design new proteins with specific functions for industrial or environmental applications.
- Fundamental Research: Understanding protein structures helps elucidate underlying biological mechanisms and discover new therapeutic targets.
Sharing via the AlphaFold database
AlphaFold has committed to sharing their technology with the research community. To this end, DeepMind has established the AlphaFold Protein Structure Database based on AlphaFold’s predictions.
This database is freely available, allowing researchers worldwide to access and use this data for their research.
It contains over 350,000 structures, including 20,000 known human proteins, as well as proteomes of other organisms significant for biological research, such as yeast and mice.
Conclusion
Thus, AlphaFold’s success in predicting protein structures illustrates the revolutionary potential of artificial intelligence and deep learning in scientific research.
To learn more about deep learning technologies and training for careers in Data Science, join DataScientest.